This blog post explains a couple of workflows to connect to Big Query using R, and access Global Fishing Watch’s public data. Since you found this post, I’m going to assume that you are somewhat familiar with Global Fishing Watch and are looking to explore and analyze the data using R. If you are not, check out our Science publication Tracking the global footprint of fisheries as well as the Global Fishing Watch Open Data portal.
What is BigQuery
BigQuery is Google’s platform to store and analyze large datasets. It is very fast and well integrated to other Google products such as cloud storage, and it’s free to use for up to 1TB of data analyzed each month, and up to 10GB of data stored. As any other database, it understands SQL and the online editor is a friendly place to learn and troubleshoot your queries. Big Query understands both Legacy and Standard SQL. Unfortunately, these languages are more different that you would expect, which makes things confusing. In this post I will use Standard SQL since Big Query is slowly migrating to only using this language in the future
To start using BigQuery, you need to have a gmail account and a Big Query-enabled project. If you don’t have this yet, please follow the steps in this quickstart guide quickstart guide and come back when done.
Global Fishing Watch’s data in BigQuery
Now that you have a BigQuery-enabled project, go the BigQuery’s UI page. You should see your project in the side bar, above the Public Datasets project. Click on the small arrow next to your project name, then switch to project -> display project, and enter “global-fishing-watch” (without the quotes) on the Project ID. Click OK, and you should then have a global-fishing-watch project in your side bar. This project contains, two datasets: "gfw_public_data" and "global_footprint_of_fisheries". We will work on the latter, which contains the public data released with the Science publication. This dataset contains the following four tables:
fishing_effort: Daily Fishing Effort and Vessel Presence at 100th Degree Resolution by Flag State and Gear Type, 2012-2016fishing_effort_byvessel:Daily Fishing Effort at 10th Degree Resolution by MMSI, 2012-2016fishing_vessels: Characteristics of each vessel included in the effort datavessels: This table includes all vessels that were matched to a registry, were identified through manual review or web searchers, or were classified by the neural network.
Establishing a connection
The first step to use the data from R, is to set a connection with BigQuery. This is done using the DBI and bigrquery packages:
library(DBI)
library(bigrquery)
BQ_connection <- dbConnect(bigquery(),
project = 'global-fishing-watch',
dataset = "global_footprint_of_fisheries",
billing = "ng-gfw", # your billing account name
use_legacy_sql = FALSE) # specify we are using Standard SQLWe then need to authenticate the connection. This can be done when running your first query such as listing the tables in your connection through the function dbListTable(). This will trigger a prompt in your R session asking if you want to cache your credentials. Allow access and return to RStudio.
DBI::dbListTables(BQ_connection)## [1] "fishing_effort" "fishing_effort_byvessel"
## [3] "fishing_vessels" "vessels"Now we are all set to start querying and analyzing Global Fishing Watch’s data. There are a couple of approaches to do this and whichever you use depends primarily on: 1) your experience with SQL, 2) how much you dislike SQL, and 3) how much you use R notebooks as opposed to classic R scripts. To illustrate these approches, I will walk through 3 simple analyses that exemplify the type of queries you may want to do:
- Summarize the number of fishing vessels by flag state
- Make a time series of fishing effort for China’s trawlers fleet
- Make a map of fishing effort for a particular region of the ocean
The first example shows how to query data without writing a single SQL statement by leveraging the power of the dbplyr package. While very convenient, this approach has some limitations that I will mention below. The second and third examples use workflows that require you to write SQL, but give you more control and flexibility to do more interesting and complex queries. These, in my experience, are the best ways to unleash the full potential of Google Big Query and Global Fishing Watch’s data and I highly recommend you to explore them.
1. Summarize the number of vessels by flag state and gear type
This is possibly one the first and simplest queries to do. For this use case, we will use the dbplyr and bigrquery packages that allow us to avoid the need to write SQL. We just write the dplyr verbs and functions we all love, and the dbplyr package translates them into SQL in the back-end. The main advantages of this approach is that it’s very friendly and readable, and prevents us from the cognitive overhead of switching between programming languages. However, SQL is a very large language and dbplyr does not do everything; it focuses on SELECT statements which are often what we use the most.
The first step here is to connect to the table we want to query using the tbl() function:
library(tidyverse) #loads dplyr and friends
fishing_vessels <- dplyr::tbl(BQ_connection,
"fishing_vessels") # connects to a tableHowever, notice that the variable created in your environment is not a dataframe or tibble but instead a list. This is because tbl() creates a reference to the table in the remote database but does not bring the actual data into memory. When you print it out, you’ll see it looks like a tibble but the class is tbl_sql.
fishing_vessels## # Source: table<fishing_vessels> [?? x 11]
## # Database: BigQueryConnection
## mmsi flag geartype length tonnage engine_power active_2012
## <int> <chr> <chr> <dbl> <dbl> <dbl> <lgl>
## 1 6.03e8 AGO trawlers 32.8 299. 734. FALSE
## 2 6.03e8 AGO trawlers 34.6 396. 865. FALSE
## 3 6.03e8 AGO trawlers 28.8 264. 652. FALSE
## 4 6.03e8 AGO trawlers 30.7 300. 704. FALSE
## 5 6.03e8 AGO trawlers 37.5 406. 851. FALSE
## 6 6.03e8 AGO trawlers 27.4 277. 749. FALSE
## 7 6.03e8 AGO trawlers 32.4 442. 889. FALSE
## 8 6.03e8 AGO trawlers 37.8 444. 887. FALSE
## 9 6.03e8 AGO trawlers 32.4 396. 812. FALSE
## 10 6.04e8 AGO purse_s… 36.8 292. 1182. TRUE
## # ... with more rows, and 4 more variables: active_2013 <lgl>,
## # active_2014 <lgl>, active_2015 <lgl>, active_2016 <lgl>class(fishing_vessels)## [1] "tbl_dbi" "tbl_sql" "tbl_lazy" "tbl"The most important difference between a local dataframe and a remote dataframe of class tbl_sql is that in the latter your R code will run directly the database, not in memory. To do this efficiently, dplyr will be as lazy as possible by 1) not pulling data into R unless explicitly asked to, and 2) delaying the actual communication to the database until the last possible moment (i.e., after it collects all that you want to do).
Now, we can continue to use the dplyr verbs we are familiar with and summarize the table as we wish:
summary_by_country_and_gear <- fishing_vessels %>%
filter(active_2016) %>%
group_by(flag, geartype) %>%
summarize(n_vessels = n_distinct(mmsi),
total_GT = sum(tonnage)) %>%
arrange(desc(n_vessels))Again, the object created in our environment is not the dataframe we expected. In fact, the above code never touched the database, it only recorded the instructions to query the data. To pull the data into a local tibble we need to explicitly ask for it with the function collect().
summary_by_country_and_gear <- collect(summary_by_country_and_gear)Now we have an actual dataframe with which we can make a simple summarizing plot:
top_20_flags <- summary_by_country_and_gear %>%
group_by(flag) %>%
summarize(n_vessels = sum(n_vessels)) %>%
top_n(20, n_vessels) %>%
pull(flag)
summary_by_country_and_gear %>%
filter(flag %in% top_20_flags) %>%
na.omit() %>%
ggplot(aes(x = forcats::fct_reorder(flag, n_vessels), y = n_vessels, fill = geartype))+
geom_col()+
coord_flip()+
labs(x = "")+
hrbrthemes::theme_ipsum()+
ggsci::scale_fill_startrek()
Chinese Trawlers are, by far, the largest fleet in the Global Fishing Watch Database.
This approach is straight forward and works perfectly for simple queries. However, I recommend you become proficient with SQL to make the most of the power of BigQuery and Global Fishing Watch data. This will help you troubleshoot when things don’t work as expected and it will make it easier to reach out to the Global Fishing Watch community for help. A couple of great resources to learn are: Learn SQL | Codecademy, and Learn SQL the Hard Way. Another way to start learning SQL is to use the function show_query() from dplyr, which will show you what your R code gets translated as in SQL, behind scenes.
fishing_vessels %>%
filter(active_2016) %>%
group_by(flag, geartype) %>%
summarize(n_vessels = n_distinct(mmsi),
total_GT = sum(tonnage)) %>%
arrange(desc(n_vessels)) %>%
show_query()## <SQL>
## SELECT `flag`, `geartype`, COUNT(DISTINCT `mmsi`) AS `n_vessels`, SUM(`tonnage`) AS `total_GT`
## FROM `fishing_vessels`
## WHERE (`active_2016`)
## GROUP BY `flag`, `geartype`
## ORDER BY `n_vessels` DESC2. Make a time series of fishing effort for China’s trawlers fleet
The dplyr way
For this analysis we need to summarize fishing by date, only for Chinese trawlers between 2012-2016. So, we need to join the effort and vessels characteristics data and filter effort for Chinese trawlers only. An inner join is the perfect operation for this.
Using the previous approach, we would write something like this:
fishing_vessels <- dplyr::tbl(BQ_connection, "fishing_vessels")
fishing_effort_byvessel <- dplyr::tbl(BQ_connection, "fishing_effort_byvessel")
ts_china_effort <- fishing_effort_byvessel %>%
inner_join(fishing_vessels %>%
filter(flag == "CHN", geartype == "trawlers"), by = "mmsi") %>%
group_by(date) %>%
summarize(fishing_hours = sum(fishing_hours, na.rm = T)) %>%
ungroup() %>%
arrange(date) %>%
collect()
head(ts_china_effort)## # A tibble: 6 x 2
## date fishing_hours
## <chr> <dbl>
## 1 2012-01-06 11.0
## 2 2012-01-07 20.6
## 3 2012-01-08 10.7
## 4 2012-01-09 7.53
## 5 2012-01-10 4.36
## 6 2012-01-11 17.4Until recently, joining datasets through bigrquery in this way was not possible. This has been implemented a couple weeks ago in the development version of the bigrquery package, so make sure to install it for this to work.
Why would I ever write SQL?
So, if I can simply write dplyr verbs to analyze data, why ever bother with SQL? Well, while the bigrquery library is a fantastic tool for simple queries, I have found some important limitations that are a deal breaker for me. Here my reasons for writing SQL directly:
- The translation of functions across languages does not always work as expected. This results is a lot of head scratching and hacky workarounds that are not intuitive. For instance, try filtering the above time series for a particular year of interest.
- If you are new to SQL, Big Query’s online editor is a fantastic learning tool. 1) It verifies your queries and tells you if/where you have a mistake. 2) It has function, variable, and table auto completion. And 3) it has tooltip helpers that describe the functions you are trying to use.
- Big Query also tells you how much data you are going to query and therefore how much are you going to be billed for. This is important - especially if you are learning - because you do not want to mistakenly write an expensive query that burns all your Google Credits. I learned this the hard way. As a rule of thumb, querying 1TB of data is equal to $6.
- Lastly, writing and understanding SQL will help you troubleshoot and get help from the GFW community. Those guys do not work in R, so if you want their help, you need to speak their language :)
OK. So hopefully I have convinced you that learning SQL is worth it. Again, some great resources to learn are: Learn SQL | Codecademy, and Learn SQL the Hard Way.
The SQL way
For the rest of this tutorial I’m going to assume that you have put in your hours and you are familiar with SQL. So, I will only focus on showing a couple workflows to write SQL and connect to Big Query without ever leaving R.
One approach is writing your queries as strings, and later using the bigrquery package to execute them using the same connection we have specified above. A great way to write your queries as strings is using the glue_sql() function from the glue library. This allows you to 1) use local R variables inside your SQL queries, and 2) divide long queries into sub-queries. This is illustrated below:
# Define your variables
min_year <- 2012
gear <- "trawlers"
flag <- "CHN"
# Query to get effort by mmsi and date after 2012
all_effort_by_mmsi_query <- glue::glue_sql(
'
Select
date,
mmsi,
fishing_hours
FROM
`global-fishing-watch.global_footprint_of_fisheries.fishing_effort_byvessel`
WHERE
EXTRACT(YEAR FROM PARSE_DATE("%F",date)) > {min_year}
',
.con = BQ_connection
)
# Query to extract only chinese trawlers
chinese_vessels_sub_query <- glue::glue_sql(
"
Select
mmsi
FROM
`global-fishing-watch.global_footprint_of_fisheries.fishing_vessels`
WHERE flag = {flag}
AND geartype = {gear}
",
.con = BQ_connection
)In each of these sub queries we are using locally defined variables by wrapping them with {}. Next, we can combine these sub-queries into a larger final query by wrapping the sub-query variable names with {} as we did above.
full_sql <- glue::glue_sql(
"
SELECT
date,
sum(fishing_hours) AS fishing_hours
FROM ({all_effort_by_mmsi_query}) AS a
LEFT JOIN ({chinese_vessels_sub_query}) AS b
ON a.mmsi = b.mmsi
GROUP BY date
ORDER BY date
DESC
",
.con = BQ_connection
)Notice that if you print the variable full_sql you can see the entire SQL query that has been built.
full_sql## <SQL> SELECT
## date,
## sum(fishing_hours) AS fishing_hours
## FROM (Select
## date,
## mmsi,
## fishing_hours
## FROM
## `global-fishing-watch.global_footprint_of_fisheries.fishing_effort_byvessel`
## WHERE
## EXTRACT(YEAR FROM PARSE_DATE("%F",date)) > 2012) AS a
## LEFT JOIN (Select
## mmsi
## FROM
## `global-fishing-watch.global_footprint_of_fisheries.fishing_vessels`
## WHERE flag = 'CHN'
## AND geartype = 'trawlers') AS b
## ON a.mmsi = b.mmsi
## GROUP BY date
## ORDER BY date
## DESCUp until this point we have not touched the actual data, we have only laid out the recipe for querying it. To actually run the query we then can use the function dbGetQuery() from the DBI library like:
china_trawlers_ts <- dbGetQuery(BQ_connection,
full_sql)You do not have to use the glue library to compose queries and execute them via this method. You can simply write the SQL queries as strings. I just wanted to show this handy function because writing long SQL queries in a single string often gets too crazy to manage without splitting things up.
The resulting object from this query is a dataframe of daily total fishing effort by Chinese trawlers, from 2013 onwards, arranged by date. Now lets plot the data to observe the effect of the Chinese new year and the summer fishing moratorium on China’s temporal patterns of fishing effort.
moratoria_dates <- tibble(year = c(2013:2016)) %>%
mutate(start_date = lubridate::ymd(paste(year,"-06-01",sep = "")),
end_date = lubridate::ymd(paste(year,"-08-01",sep = "")))
new_year_dates <- tibble(year = c(2013:2016),
start_date = c(lubridate::ymd("2013-02-07"),
lubridate::ymd("2014-01-28"),
lubridate::ymd("2015-02-16"),
lubridate::ymd("2016-02-05")),
end_date = c(lubridate::ymd("2013-02-13"),
lubridate::ymd("2014-02-3"),
lubridate::ymd("2015-02-22"),
lubridate::ymd("2016-02-11")))ggplot() +
geom_rect(data = moratoria_dates,
aes(xmin = start_date,
xmax = end_date,
ymin = 0,
ymax = Inf,
fill = "navyblue"),
alpha = 0.5,
show.legend = TRUE) +
geom_rect(data = new_year_dates,
aes(xmin = start_date,
xmax = end_date,
ymin = 0,
ymax = Inf,
fill = "dodgerblue"),
alpha = 0.5,
show.legend = TRUE) +
geom_line(data = china_trawlers_ts %>%
mutate(date = lubridate::ymd(date)) %>%
filter(lubridate::year(date) > 2012),
aes(x = date, y = fishing_hours),
size = 0.3)+
theme_minimal() +
theme(axis.ticks = element_line(size = 0.5),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
axis.text.y = element_text(size = 10),
axis.text.x = element_text(size = 10),
axis.title = element_text(size = 10),
legend.text = element_text(size = 6),
legend.justification = "center",
legend.position = "bottom",
plot.margin = margin(2,2,2,2)) +
scale_x_date(date_breaks = "1 year",
date_labels = "%Y ") +
xlab("") +
scale_y_continuous(expand = c(0, 0),
labels = scales::comma) +
ylab("Fishing hours")+
scale_fill_manual(values = c("orange", "dodgerblue"),
name = " ",
labels = c("Chinese New Year", "Moratoria")) +
guides(colour = guide_legend(override.aes = list(alpha = .5)))
Important conclusion: Holidays can have as large an effect as strictly enforced policies on fishing activity. We need more holidays!
3. Make a map of fishing effort for a particular region of the ocean
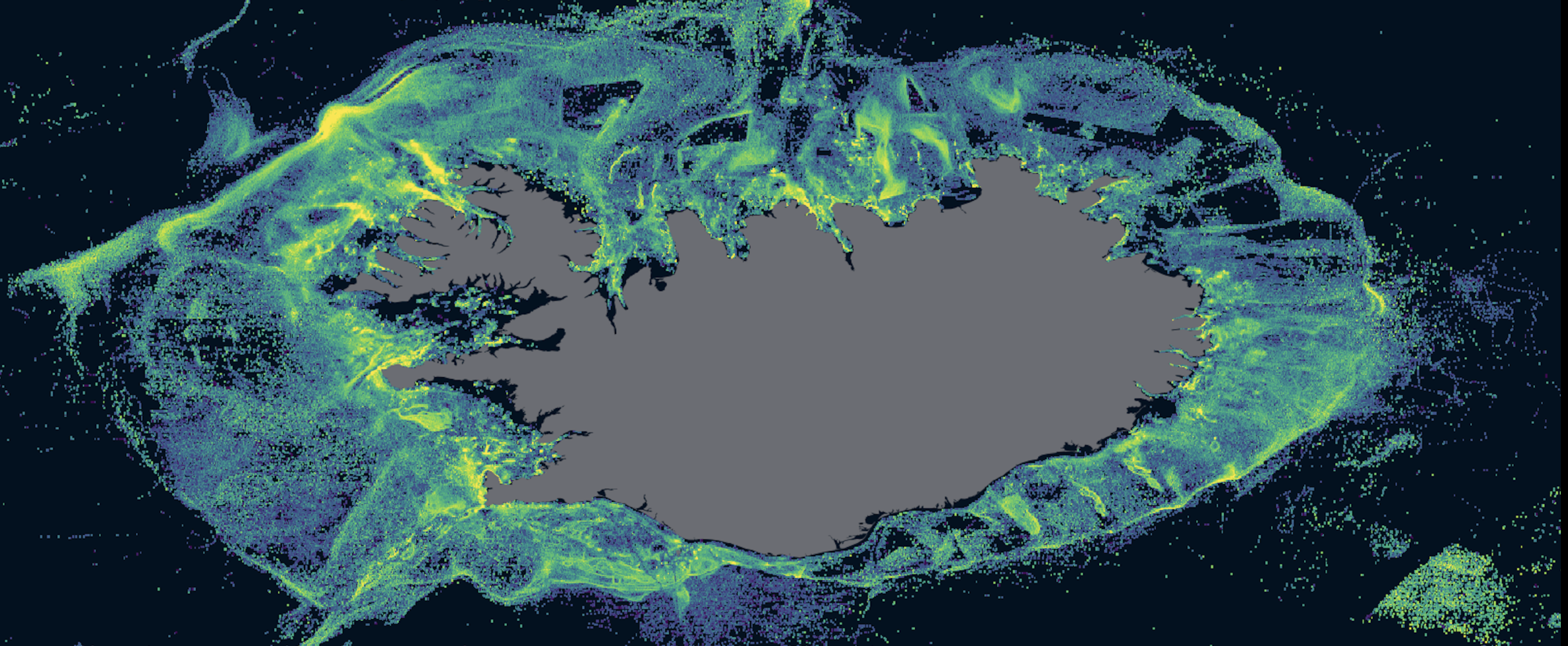
A common use of Global Fishing Watch’s data is to make maps of fishing effort in an area of interest. I will show how you can do this and highlight another approach to query data from within R, using R Notebooks and chunks. For this, let’s make a map of fishing effort around the Azores Archipelago, Portugal.
If you are not familiar with R Notebooks, they are a type of code script that allows you to mix prose and code in a single document, with chunks of code that can be run independently and interactively. This is called literate programming, and makes analysis easier to reproduce. In fact, this tutorial is an R Notebook!. For more details on R Notebooks take a look at R Markdown: The Definitive Guide by Yihui Xie. One important advantage of R Notebooks is that chunks of code can use pretty much any programming language. This means that you could mix python, SQL, and R in a single, easy to read document! For our example we will use a SQL chunk to directly write an SQL query and get the data of fishing effort around the Azores.
First, lets get a map of the Azores to obtain the bounding of of our region of interest. To do this we can use the mregions package and a couple handy functions from the sf package.
# get the shapefile from mregions
azores_eez <- mregions::mr_shp(key = "MarineRegions:eez",
filter = "Portuguese Exclusive Economic Zone (Azores)",
maxFeatures = 200)
# transform the shapefile into an sf object
azores_eez <- azores_eez %>%
sf::st_as_sf()
# get the bounding box of the shapefile
azores_bbox <- sf::st_bbox(azores_eez)
# extend the bounding box 1 degree in every direction.
min_lon <- azores_bbox[["xmin"]] - 1
max_lon <- azores_bbox[["xmax"]] + 1
min_lat <- azores_bbox[["ymin"]] - 1
max_lat <- azores_bbox[["ymax"]] + 1
# define mapping resoluion in degrees
resolution <- 0.1Now, we query the data. To do this with a sql chunk, we need to simply specify the language we are using (sql), the database connection (same as before), and the name we want to give to the variable that will contain the results of the query.
`
``{sql connection = BQ_connection, output.var = "binned_effort_around_Azores", echo = FALSE}
SELECT
FLOOR(lat_bin/?resolution)*?resolution + 0.5*?resolution lat_bin_center,
FLOOR(lon_bin/?resolution)*?resolution + 0.5*?resolution lon_bin_center,
SUM(fishing_hours) fishing_hours
FROM (
SELECT
lat_bin/100 lat_bin,
lon_bin/100 lon_bin,
fishing_hours
FROM
`global-fishing-watch.global_footprint_of_fisheries.fishing_effort`
WHERE
_PARTITIONTIME >= '2016-01-01 00:00:00'
AND _PARTITIONTIME < '2016-12-31 00:00:00')
WHERE
lat_bin >= ?min_lat
AND lat_bin <= ?max_lat
AND lon_bin >= ?min_lon
AND lon_bin <= ?max_lon
GROUP BY
lat_bin_center,
lon_bin_center
HAVING
fishing_hours > 0
```OK, so what just happened here? First, we filtered the data for the year 2016 and converted the coordinates from 100th of a degree to degrees (See table schema in BigQuery). Then we filtered the coordinates so that they are inside our bounding box. To do this we add ? before the name of our locally defined variable and its value gets automagically added to the query. Lastly, we group and summarize the effort data with a resolution of 0.1º degrees and exclude anything with no fishing effort.
binned_effort_around_Azores %>%
filter(fishing_hours > 1) %>%
ungroup() %>%
ggplot()+
geom_raster(aes(x = lon_bin_center, y = lat_bin_center, fill = fishing_hours))+
viridis::scale_fill_viridis(name = "Fishing hours" ,
trans = "log",
breaks = scales::log_breaks(n = 10, base = 4))+
geom_sf(data = azores_eez,
fill = NA,
col = "black",
size = 1.4
) +
hrbrthemes::theme_ipsum()+
labs(title = "Fishing effort inside and around Azores",
subtitle = "2017",
y = "",
x = "")+
theme(axis.text.x = element_text(size = 10),
axis.text.y = element_text(size = 10),
plot.title = element_text(size = 14, hjust = 0),
plot.subtitle = element_text(size = 14, hjust = 0),
plot.margin = margin(10,0,0,0))
Closing remarks
Global Fishing Watch’s commitment to transparency and open source data, is rapidly changing the way we do research in fisheries science, and marine conservation. Powerful tools like Big Query and useful R packages (e.g., DBI, bigrquery, rmarkdown) are making these data accessible and easy to analyze, which will hopefully increase the rate it’s being used and the impact it can have. Have fun!